publications
Peer-reviewed research articles on microbiome analysis, machine learning, and microbial interactions
2024
-
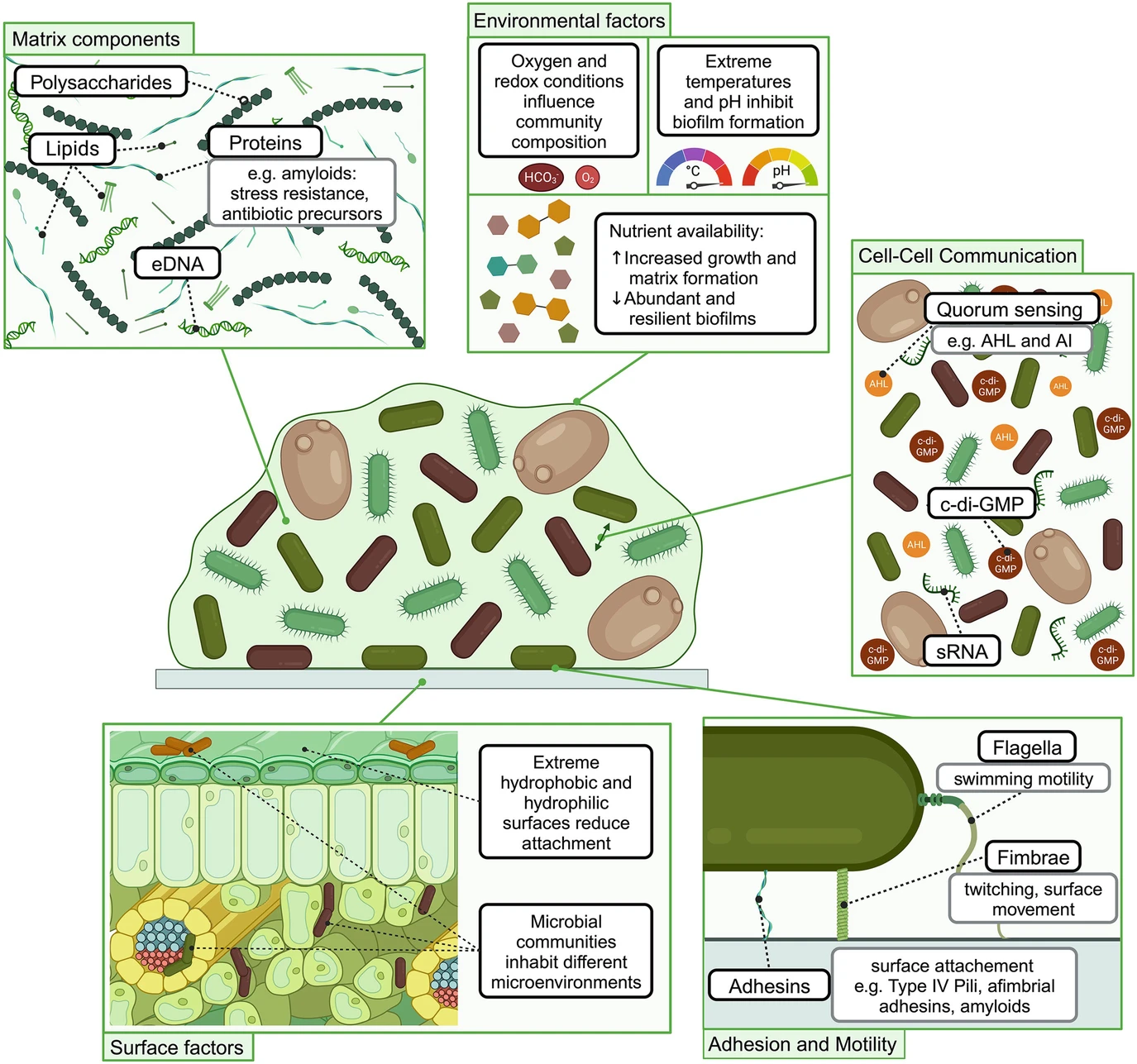 Nature’s Protectors: A Biofilm Perspective on Bacterial Disease Control in PlantsDaniel Gómez-Pérez, Leonie M. Zott, Monja Schmid, and 1 more authorPlant-Microbe Interaction and Stress Management, 2024
Nature’s Protectors: A Biofilm Perspective on Bacterial Disease Control in PlantsDaniel Gómez-Pérez, Leonie M. Zott, Monja Schmid, and 1 more authorPlant-Microbe Interaction and Stress Management, 2024Bacterial diseases significantly threaten agricultural productivity, necessitating innovative and sustainable approaches to disease management. Above- and belowground biofilms are essential in mitigating plant bacterial diseases. We examine aboveground biofilms, primarily located on leaf surfaces, for their ability to form protective barriers and produce antimicrobial compounds, thereby impeding the establishment of pathogenic bacteria. Concurrently, we explore belowground biofilms in the rhizosphere for their contributions to nutrient cycling, enhanced nutrient uptake, and the deployment of biocontrol agents against soilborne bacterial pathogens. Here, we employ a multidisciplinary approach by integrating molecular, microbiological, and ecological analyses to unravel the mechanisms underlying the formation and function of these beneficial biofilms. We investigate quorum sensing, microbial communication, and the intricate interplay between plant hosts and beneficial microbes to elucidate the complex networks orchestrating disease resistance. We consider environmental factors such as surface structure, temperature, and nutrient availability. We acknowledge their impact on biofilm-mediated disease management strategies relevant for controlling plant disease. Furthermore, we emphasize the importance of microbial diversity within biofilms for its role in enhancing plant resilience to bacterial diseases. Overall, this chapter provides a foundation for developing targeted and sustainable strategies in bacterial disease management, underpinning the critical role of above- and belowground beneficial biofilms. Insights gained from beneficial biofilms contribute to the fundamental understanding of synergistic relationships in the plant microbiome environment and their implications for sustainable agriculture
@article{Gómez-Pérez.2024, year = {2024}, title = {{Nature’s Protectors: A Biofilm Perspective on Bacterial Disease Control in Plants}}, author = {Gómez-Pérez, Daniel and Zott, Leonie M. and Schmid, Monja and Chaudhry, Vasvi}, journal = {Plant-Microbe Interaction and Stress Management}, issn = {2523-8442}, doi = {10.1007/978-981-97-4239-4\_7}, pages = {109--133}, keywords = {}, dimensions = {true}, }
2023
-
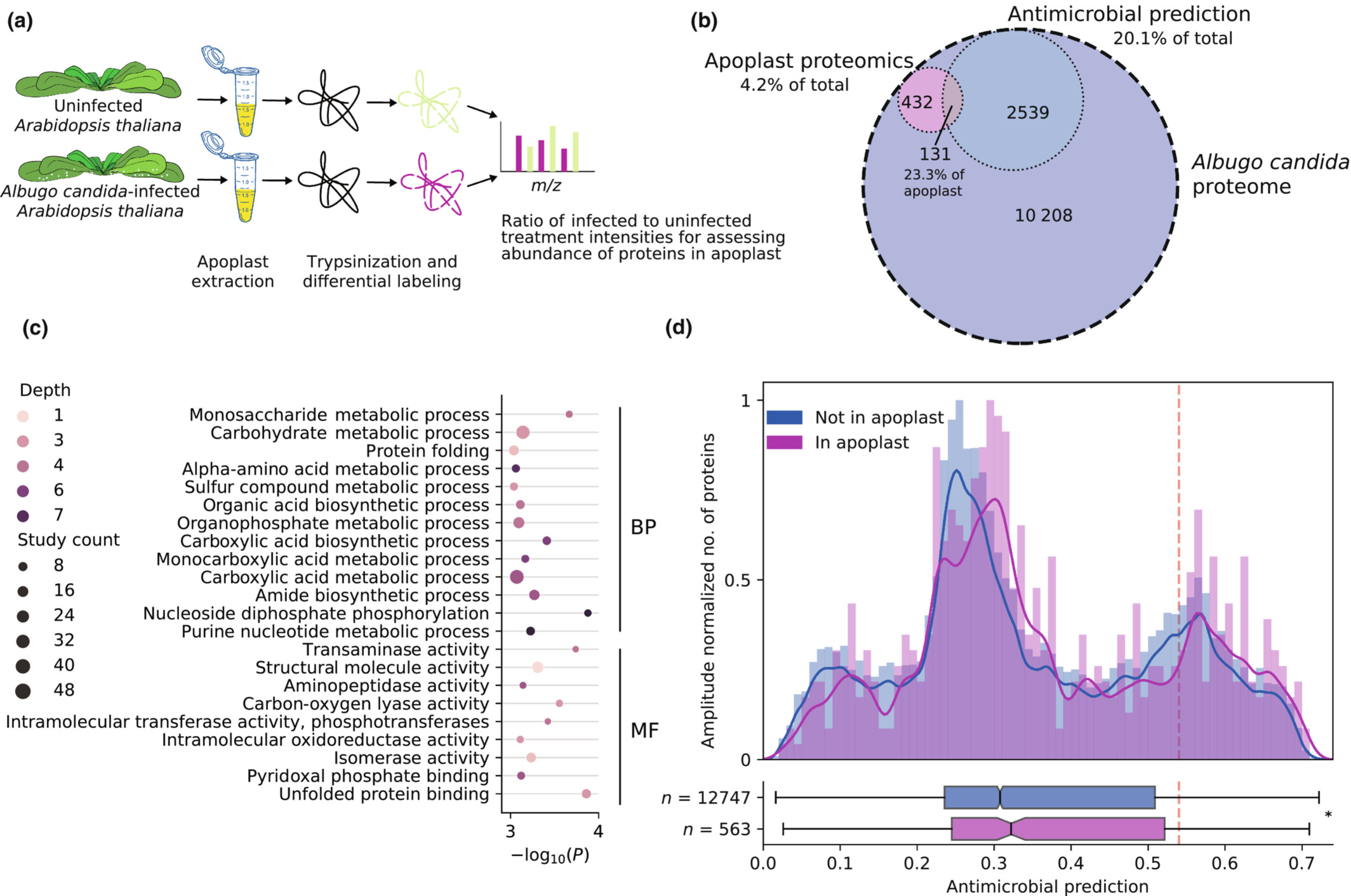 Proteins released into the plant apoplast by the obligate parasitic protist Albugo selectively repress phyllosphere-associated bacteriaDaniel Gómez-Pérez, Monja Schmid, Vasvi Chaudhry, and 6 more authorsNew Phytologist, 2023
Proteins released into the plant apoplast by the obligate parasitic protist Albugo selectively repress phyllosphere-associated bacteriaDaniel Gómez-Pérez, Monja Schmid, Vasvi Chaudhry, and 6 more authorsNew Phytologist, 2023Biotic and abiotic interactions shape natural microbial communities. The mechanisms behind microbe–microbe interactions, particularly those protein based, are not well understood. We hypothesize that released proteins with antimicrobial activity are a powerful and highly specific toolset to shape and defend plant niches. We have studied Albugo candida, an obligate plant parasite from the protist Oomycota phylum, for its potential to modulate the growth of bacteria through release of antimicrobial proteins into the apoplast. Amplicon sequencing and network analysis of Albugo-infected and uninfected wild Arabidopsis thaliana samples revealed an abundance of negative correlations between Albugo and other phyllosphere microbes. Analysis of the apoplastic proteome of Albugo-colonized leaves combined with machine learning predictors enabled the selection of antimicrobial candidates for heterologous expression and study of their inhibitory function. We found for three candidate proteins selective antimicrobial activity against Gram-positive bacteria isolated from A. thaliana and demonstrate that these inhibited bacteria are precisely important for the stability of the community structure. We could ascribe the antibacterial activity of the candidates to intrinsically disordered regions and positively correlate it with their net charge. This is the first report of protist proteins with antimicrobial activity under apoplastic conditions that therefore are potential biocontrol tools for targeted manipulation of the microbiome.
@article{gomez-perez2023, author = {Gómez-Pérez, Daniel and Schmid, Monja and Chaudhry, Vasvi and Hu, Yiheng and Velic, Ana and Maček, Boris and Ruhe, Jonas and Kemen, Ariane and Kemen, Eric}, year = {2023}, title = {Proteins released into the plant apoplast by the obligate parasitic protist <i>Albugo</i> selectively repress phyllosphere-associated bacteria}, journal = {New Phytologist}, dimensions = {true}, volume = {230}, number = {6}, pages = {2320-2334}, keywords = {antimicrobial proteins, apoplastic proteins, intrinsically disordered proteins, microbe–microbe interactions, microbial community structure, microbiome, phyllosphere microbes, plant pathogen}, doi = {10.1111/nph.18995}, url = {https://doi.org/10.1111/nph.18995}, } -
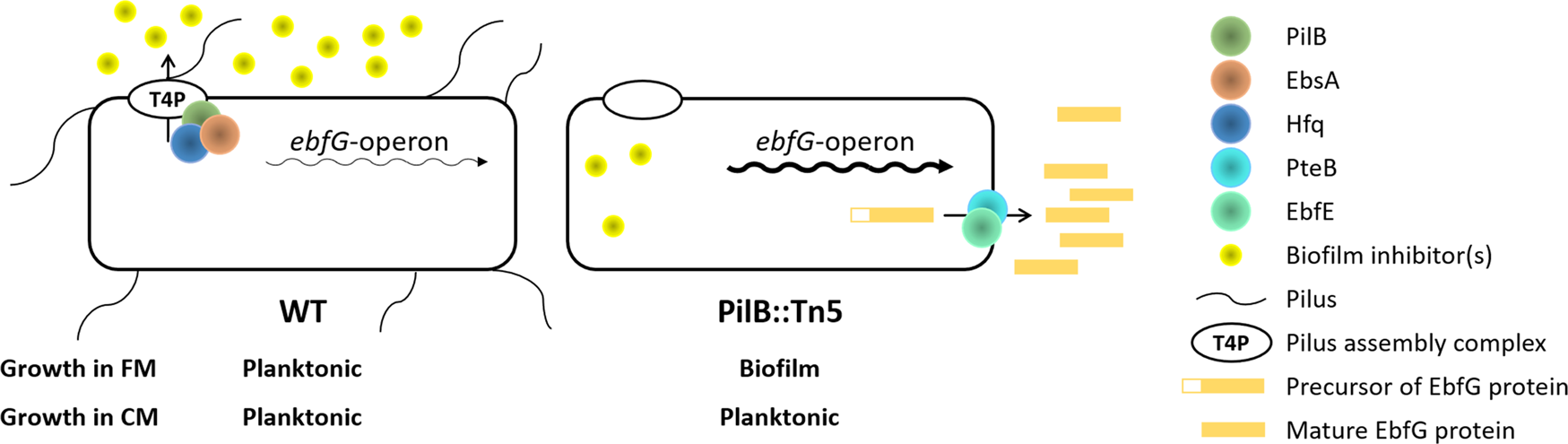 Cell specialization in cyanobacterial biofilm development revealed by expression of a cell-surface and extracellular matrix proteinAlona Frenkel, Eli Zecharia, Daniel Gómez-Pérez, and 9 more authorsnpj Biofilms and Microbiomes, 2023
Cell specialization in cyanobacterial biofilm development revealed by expression of a cell-surface and extracellular matrix proteinAlona Frenkel, Eli Zecharia, Daniel Gómez-Pérez, and 9 more authorsnpj Biofilms and Microbiomes, 2023Cyanobacterial biofilms are ubiquitous and play important roles in diverse environments, yet, understanding of the processes underlying the development of these aggregates is just emerging. Here we report cell specialization in formation of Synechococcus elongatus PCC 7942 biofilms—a hitherto unknown characteristic of cyanobacterial social behavior. We show that only a quarter of the cell population expresses at high levels the four-gene ebfG-operon that is required for biofilm formation. Almost all cells, however, are assembled in the biofilm. Detailed characterization of EbfG4 encoded by this operon revealed cell-surface localization as well as its presence in the biofilm matrix. Moreover, EbfG1-3 were shown to form amyloid structures such as fibrils and are thus likely to contribute to the matrix structure. These data suggest a beneficial ‘division of labor’ during biofilm formation where only some of the cells allocate resources to produce matrix proteins—‘public goods’ that support robust biofilm development by the majority of the cells. In addition, previous studies revealed the operation of a self-suppression mechanism that depends on an extracellular inhibitor, which supresses transcription of the ebfG-operon. Here we revealed inhibitor activity at an early growth stage and its gradual accumulation along the exponential growth phase in correlation with cell density. Data, however, do not support a threshold-like phenomenon known for quorum-sensing in heterotrophs. Together, data presented here demonstrate cell specialization and imply density-dependent regulation thereby providing deep insights into cyanobacterial communal behavior.
@article{frenkel2023, author = {Frenkel, Alona and Zecharia, Eli and G{\'o}mez-P{\'e}rez, Daniel and Sendersky, Eleonora and Yegorov, Yevgeni and Jacob, Avi and Benichou, Jennifer I. C. and Stierhof, York-Dieter and Parnasa, Rami and Golden, Susan S. and Kemen, Eric and Schwarz, Rakefet}, date = {2023/03/02}, date-added = {2023-06-04 21:34:02 +0200}, date-modified = {2023-06-04 21:34:02 +0200}, doi = {10.1038/s41522-023-00376-6}, id = {Frenkel2023}, isbn = {2055-5008}, journal = {npj Biofilms and Microbiomes}, number = {1}, pages = {10}, title = {Cell specialization in cyanobacterial biofilm development revealed by expression of a cell-surface and extracellular matrix protein}, url = {https://doi.org/10.1038/s41522-023-00376-6}, volume = {9}, year = {2023}, bdsk-url-1 = {https://doi.org/10.1038/s41522-023-00376-6}, dimensions = {true}, } -
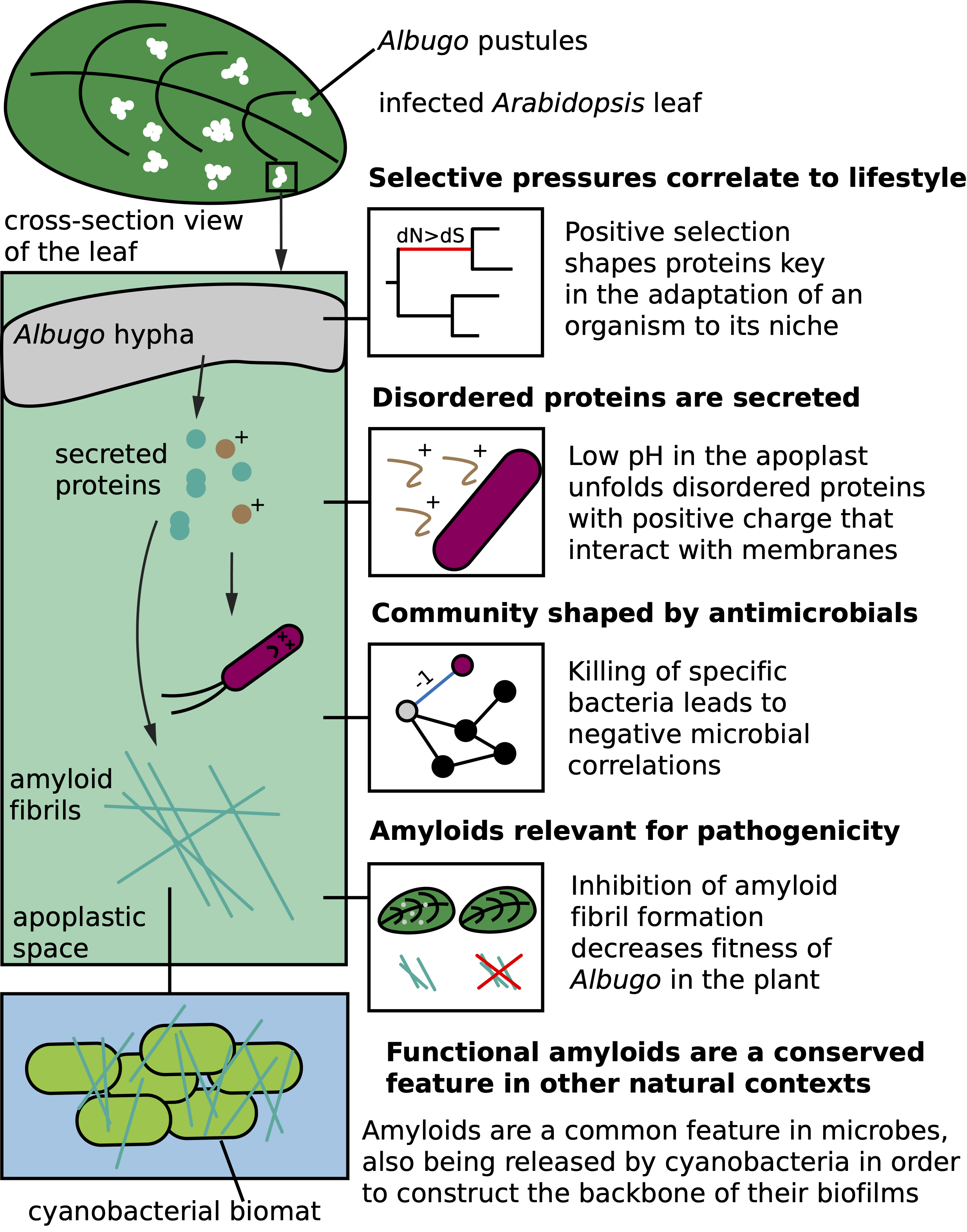 Protein-based interactions in microbial communities: the roles of amyloids and antimicrobialsDaniel Gómez-Pérez2023
Protein-based interactions in microbial communities: the roles of amyloids and antimicrobialsDaniel Gómez-Pérez2023Microbial communities ubiquitously inhabit the natural world. Members of these have evolved countless dynamic and complex relationships to ensure their survival. Whether through formation of protective enclosures, such as biofilms held together by amyloids, or actively killing competitors as antimicrobials, secreted proteins play key roles in microbial interactions. However, much remains to be understood about the specific mechanisms of activity behind many of these interactions. The oomycetal pathogen Albugo is an obligate plant biotroph that strongly modifies its surrounding microbial community. It relies on specific proteins to exert its influence, as it has lost a large part of the biosynthetic power of free-living relatives, in part due to adaptation to an obligate biotrophic lifestyle. Firstly, we have put these proteins into an evolutionary context by studying the link between lifestyle and genome features in the oomycetes. This phylum comprises Albugo as well as other plant and animal pathogens with widely divergent lifestyles and hosts. Furthermore, through a proteomics approach followed by heterologous expression, we have pinpointed as well as functionally and structurally characterized a number of proteins from Albugo that we found to be influential in controlling the surrounding microbial community. In particular, we have focused on those with antimicrobial potential as well as the ability to form amyloids. The former are interesting due to their direct role in antagonistic interactions and the high demand for novel and highly specific peptide-based antimicrobial compounds which could aid against the rise of multidrug resistant microbes. When studying antimicrobial proteins in Albugo, we could relate their effects to intrinsic disorder and high positive charge. The amyloid fold, instead, is a prevalent and overlooked characteristic of many proteins relevant to microbial interactions and to survival in particular. Because of their original discovery as etiological agents of human pathology, their study has been historically confined to the medical field. Based on current literature, however, the amyloid fold is now known to be omnipresent in the natural microbial world where it plays varied functional roles, including defense through antimicrobial activity. The protein candidates we have described in Albugo support the presence of this characteristic fold and functional relevance in protists as well, since we found amyloids to be important for pathogenicity. Finally, we have explored the amyloid-forming characteristics of proteins released by a cyanobacterium, Synechococcus elongatus, which are upregulated during the biofilm establishment stage. On the whole, we have studied and described protein-based mechanisms relevant to complex microbial communities in natural ecosystems, focusing on amyloids and antimicrobials. These highlight the countless mechanisms that could be translated to biotechnological applications and the many that are yet to be discovered.
@phdthesis{gomez-perez2023thesis, author = {G{\'o}mez-P{\'e}rez, Daniel}, copyright = {ubt-podok}, date-added = {2023-06-05 22:38:51 +0200}, date-modified = {2023-06-05 22:38:51 +0200}, doi = {10.15496/PUBLIKATION-83115}, keywords = {Mikrobiologie , Amyloid , Antimikrobieller Wirkstoff , Mikroflora, 570, molekulare Wechselwirkungen, Plant pathogen, Pflanzenpathogen, Microbiome, <i>Albugo</i>, Molecular interactions, Arabidopsis, Proteine, Amyloide, Proteins, Amyloids, Antimicrobials}, language = {en}, publisher = {Universit{\"a}t T{\"u}bingen}, title = {Protein-based interactions in microbial communities: the roles of amyloids and antimicrobials}, url = {https://publikationen.uni-tuebingen.de/xmlui/handle/10900/141768}, year = {2023}, }
2021
-
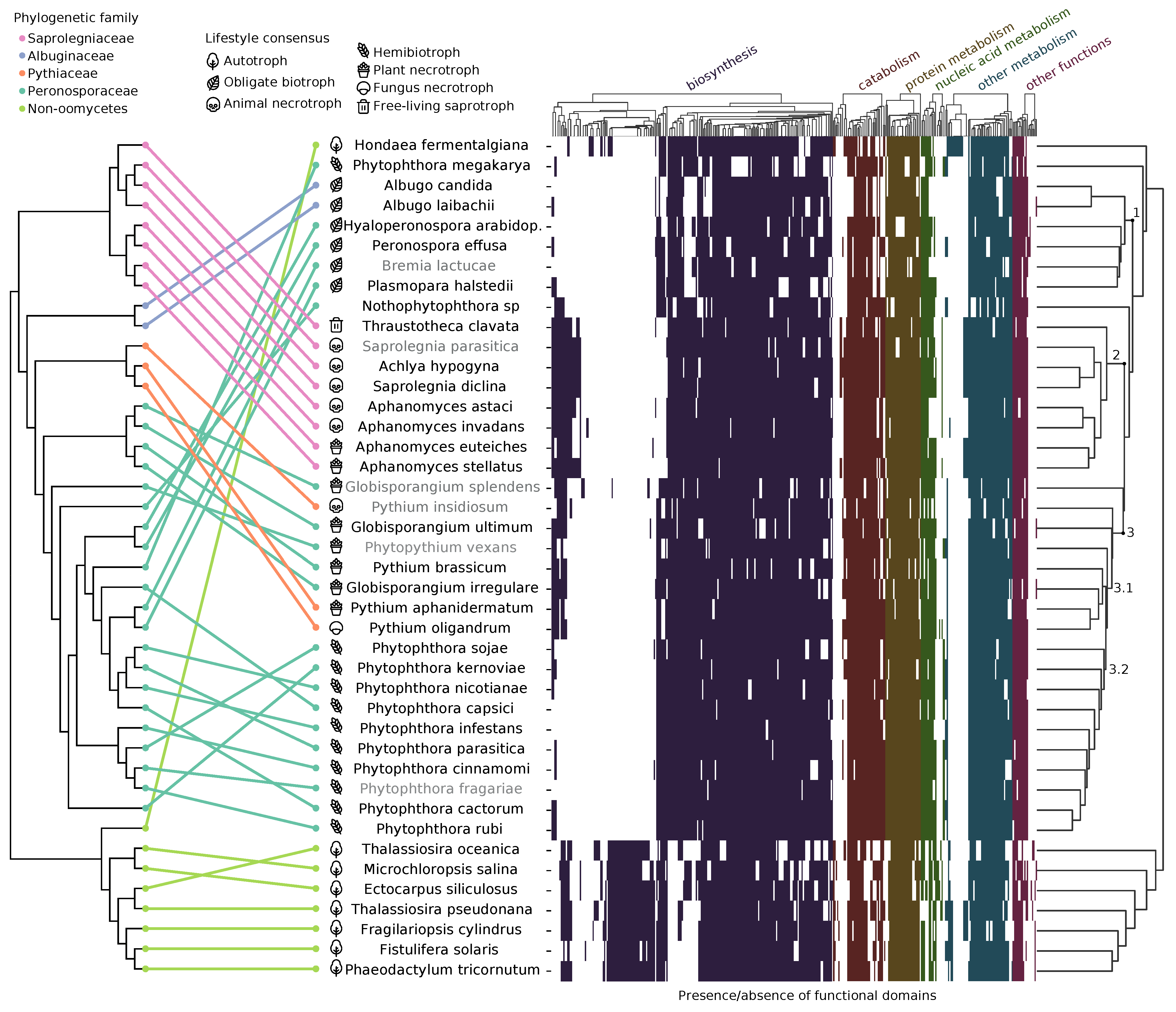 Predicting Lifestyle from Positive Selection Data and Genome Properties in OomycetesDaniel Gómez-Pérez, and Eric KemenPathogens, 2021
Predicting Lifestyle from Positive Selection Data and Genome Properties in OomycetesDaniel Gómez-Pérez, and Eric KemenPathogens, 2021As evidenced in parasitism, host and niche shifts are a source of genomic and phenotypic diversification. Exemplary is a reduction in the core metabolism as parasites adapt to a particular host, while the accessory genome often maintains a high degree of diversification. However, selective pressures acting on the genome of organisms that have undergone recent lifestyle or host changes have not been fully investigated. Here, we developed a comparative genomics approach to study underlying adaptive trends in oomycetes, a eukaryotic phylum with a wide and diverse range of economically important plant and animal parasitic lifestyles. Our analysis reveals converging evolution on biological processes for oomycetes that have similar lifestyles. Moreover, we find that certain functions, in particular carbohydrate metabolism, transport, and signaling, are important for host and environmental adaptation in oomycetes. Given the high correlation between lifestyle and genome properties in our oomycete dataset, together with the known convergent evolution of fungal and oomycete genomes, we developed a model that predicts plant pathogenic lifestyles with high accuracy based on functional annotations. These insights into how selective pressures correlate with lifestyle may be crucial to better understand host/lifestyle shifts and their impact on the genome.
@article{gomez-perez2021, author = {Gómez-Pérez, Daniel and Kemen, Eric}, title = {Predicting Lifestyle from Positive Selection Data and Genome Properties in Oomycetes}, journal = {Pathogens}, volume = {10}, year = {2021}, number = {7}, article-number = {807}, url = {https://www.mdpi.com/2076-0817/10/7/807}, pubmedid = {34202069}, issn = {2076-0817}, doi = {10.3390/pathogens10070807}, dimensions = {true}, } -
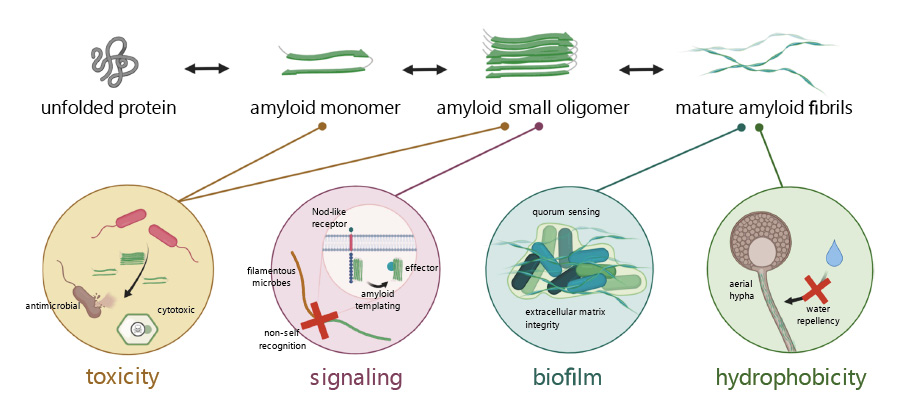 Amyloid Proteins in Plant-Associated Microbial CommunitiesDaniel Gómez-Pérez, Vasvi Chaudhry, Ariane Kemen, and 1 more authorMicrobial Physiology, Jun 2021
Amyloid Proteins in Plant-Associated Microbial CommunitiesDaniel Gómez-Pérez, Vasvi Chaudhry, Ariane Kemen, and 1 more authorMicrobial Physiology, Jun 2021Amyloids have proven to be a widespread phenomenon rather than an exception. Many proteins presenting the hallmarks of this characteristic beta sheet-rich folding have been described to date. Particularly common are functional amyloids that play an important role in the promotion of survival and pathogenicity in prokaryotes. Here, we describe important developments in amyloid protein research that relate to microbe-microbe and microbe-host interactions in the plant microbiome. Starting with biofilms, which are a broad strategy for bacterial persistence that is extremely important for plant colonization. Microbes rely on amyloid-based mechanisms to adhere and create a protective coating that shelters them from external stresses and promotes cooperation. Another strategy generally carried out by amyloids is the formation of hydrophobic surface layers. Known as hydrophobins, these proteins coat the aerial hyphae and spores of plant pathogenic fungi, as well as certain bacterial biofilms. They contribute to plant virulence through promoting dissemination and infectivity. Furthermore, antimicrobial activity is an interesting outcome of the amyloid structure that has potential application in medicine and agriculture. There are many known antimicrobial amyloids released by animals and plants; however, those produced by bacteria or fungi remain still largely unknown. Finally, we discuss amyloid proteins with a more indirect mode of action in their host interactions. These include virulence-promoting harpins, signaling transduction that functions through amyloid templating, and root nodule bacteria proteins that promote plant-microbe symbiosis. In summary, amyloids are an interesting paradigm for their many functional mechanisms linked to bacterial survival in plant-associated microbial communities.
@article{gomez-perez2021amyloid, author = {Gómez-Pérez, Daniel and Chaudhry, Vasvi and Kemen, Ariane and Kemen, Eric}, title = {{Amyloid Proteins in Plant-Associated Microbial Communities}}, journal = {Microbial Physiology}, volume = {31}, number = {2}, pages = {88-98}, year = {2021}, month = jun, issn = {2673-1665}, doi = {10.1159/000516014}, url = {https://doi.org/10.1159/000516014}, dimensions = {true}, }